James Boyko

Assistant Professor in Ecology and Evolutionary Biology at University of Michigan.
Research
There are two main themes in my research:
-
How Traits Depend on Each Other
Traits in organisms don’t evolve in isolation. A change in one trait can influence changes in another. Ignoring these connections can lead to misleading conclusions. -
Evolution Happens at Different Speeds
Not all species evolve in the same way or at the same pace. Some traits change quickly, while others stay the same for long periods. Failing to consider this uneven pace of change can also lead to incorrect inferences.
These two ideas—traits being connected and evolution happening at different speeds—are more closely linked than they might seem. This connection is particularly important in the tools we commonly use to study macroevolution, called phylogenetic comparative methods (PCMs).
Deep Learning for Evolutionary Research

The diversity of shapes and forms in living things is staggering. But can all this variety be boiled down to just a few basic patterns? If so, finding these low dimensional patterns can reveal how species have evolved over time.
I use deep-learning tools like autoencoders to identify these patterns. Autoencoders are computer models that find the most important features in complex data, like the shapes of bones or leaves. I compare these tools to more traditional methods, like principal component analysis (PCA), and find that deep learning can sometimes do a better job, especially for more complex shapes.
This work opens up exciting possibilities for improving our methods, like adding evolutionary relationships (phylogeny) or experimenting with new model designs.
Generalized Hidden Markov Models
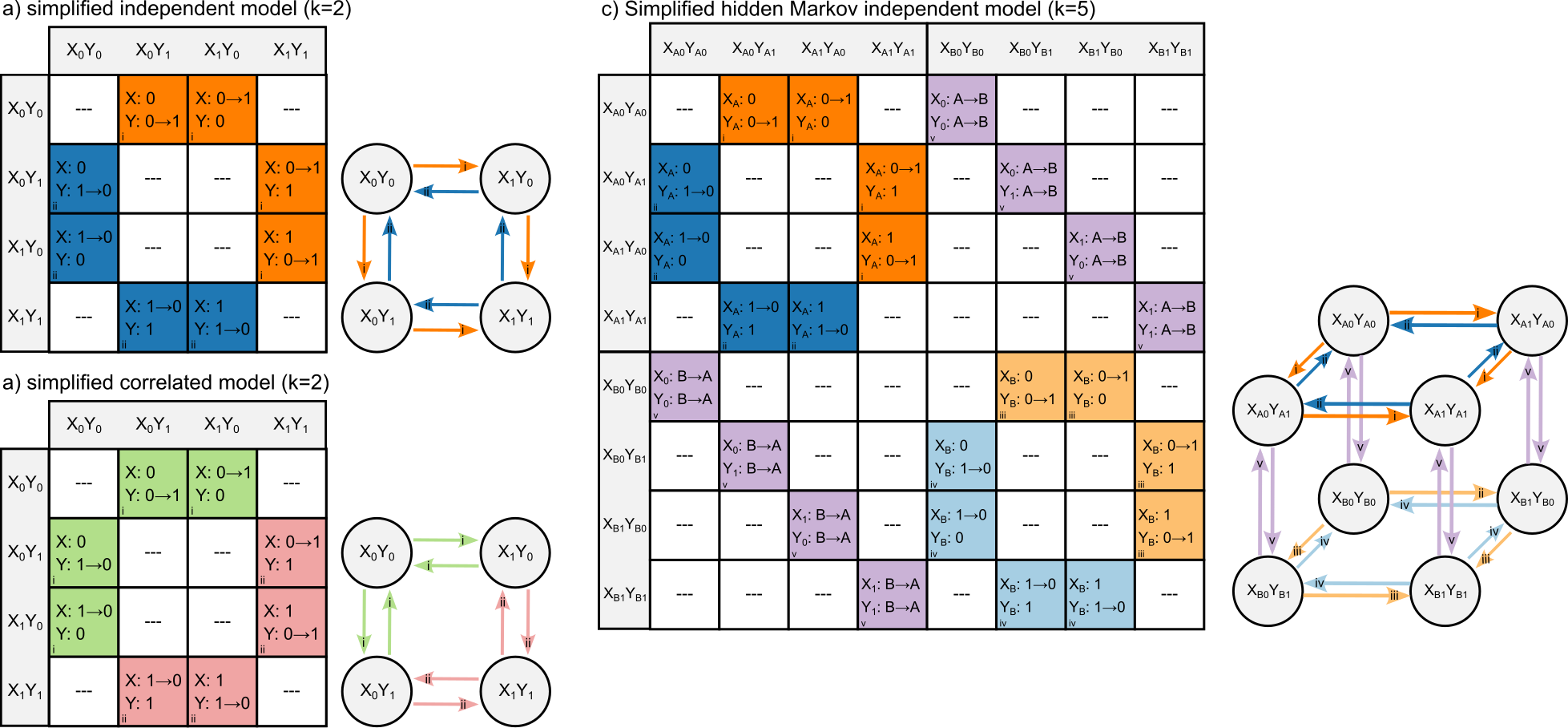
To better understand how traits evolve together, I improved existing models to handle more complex scenarios. These updated models allow us to study multiple traits at once, even when some traits are hidden or hard to observe. This approach helps us see how traits influence each other and evolve together.
Using this method, I showed that looking at traits as part of a bigger system (instead of individually) gives us more accurate insights into how species have evolved. This also helps us avoid false conclusions when reconstructing the history of traits in different species.
Linking Discrete and Continuous Traits
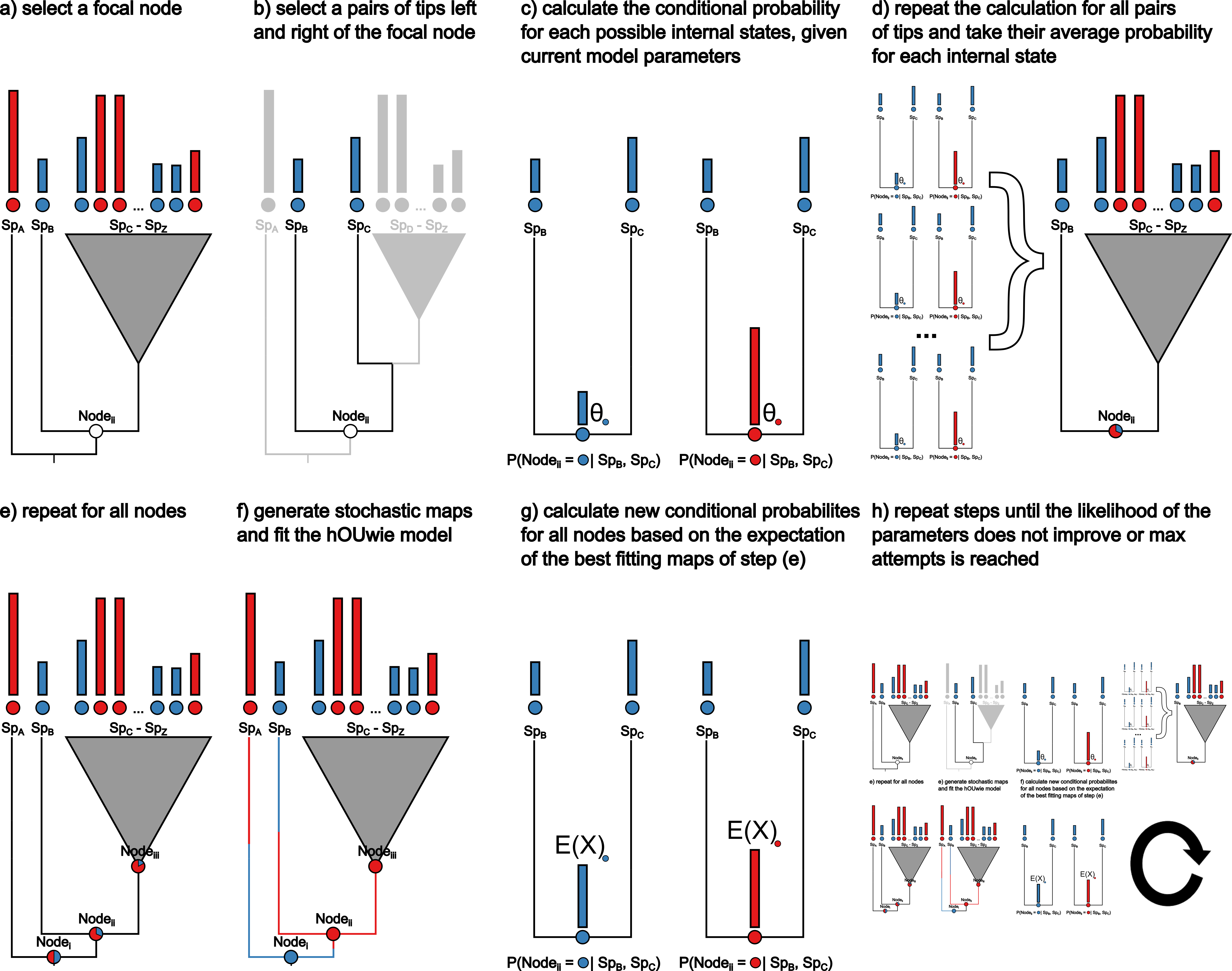
I developed a new model, hOUwie, to explore the relationship between traits and measurements, like the shape of a leaf and the climate it grows in. This model helps us understand if and how these things are connected.
A key feature of this model is that it accounts for the possibility that these connections aren’t simple or direct. It also